Signal transduction networks including ERK elicit multiple cellular functions. One of the critical properties of the signal transduction system is that the same signaling networks can code multiple cellular functions. This coding system can be generated by the combination of distinct signaling molecules/networks and by distinct temporal coding systems.
In PC12 cells, EGF and NGF induce transient and sustained ERK activation, leading to cell proliferation and differentiation, respectively. We found that transient ERK activation encode information of increasing rate of growth factors and sustained ERK activation encode that of final concentration of growth factors (Sasagawa et al., Nat. Cell Biol., 2005). We developed quantitative image cytometry (QIC), which allows us to measure signaling activities at single cell level with one minute-interval (Ozaki et al., PLoS ONE, 2010), and made a system identification of temporal decoding of ERK activation by selective immediate early genes (IEGs) expression (Saito et al., PLoS ONE, 2013). We found that pulsatile ERK phosphorylation was decoded by selective expression of EGR1 rather than c-FOS, and conjunctive NGF and PACAP stimulation was decoded by synergistic JUNB expression through a switch-like response to c-FOS.
The relationship between signaling molecules and downstream gene expression levels and cell-fate decision is a multiple-input and multiple-output (MIMO) system. We applied a statistical linear multivariate regression with reduction by variable elimination (backward elimination PLS regression) to growth factor-specific signaling and cell-fate decisions in PC12 cells, and extracted simple relationships of the MIMO system (Akimoto, et al, PLoS ONE, 2013).
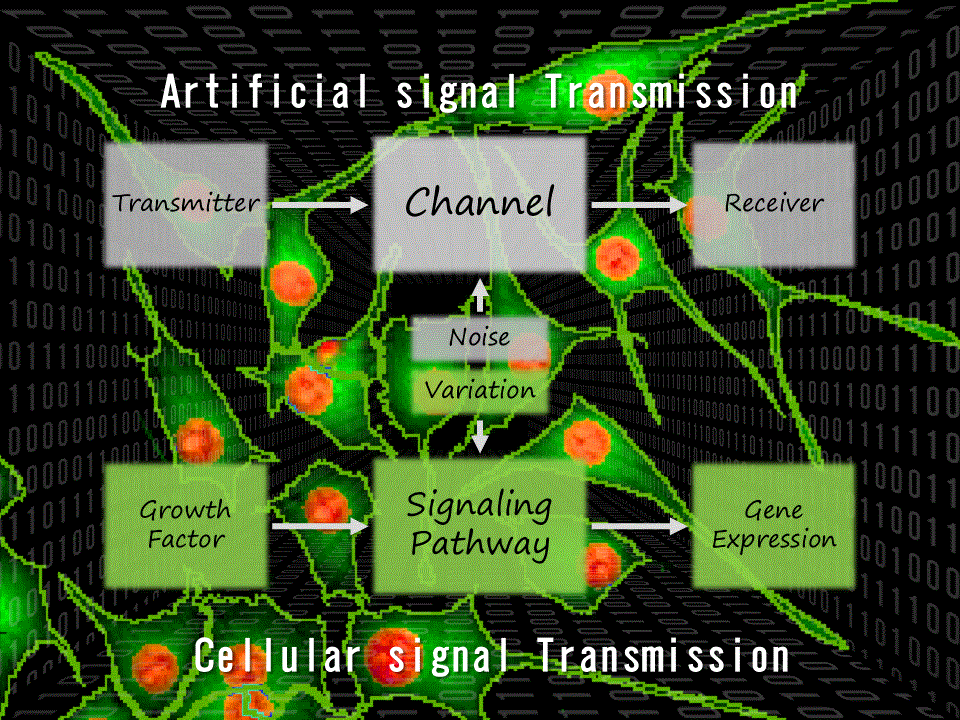
We also analyzed the signaling pathway in the framework of Shannon’s information theory, and found that information transmission was generally more robust than average signal intensity despite pharmacological perturbations, and information transmission through unperturbed signaling pathways compensatorily increased in many signaling pathways (Uda, S. et al., Science, 2013) (Fig. 3).
|
|
|